The Matrix is the world that has been pulled over your eyes to blind you from the truth.
|
HOWTO /
TimeEvolutionIn this tutorial, we will calculate the real-time evolution of a spin excitation in the S=1/2 Heisenberg model. In this tutorial we just calculate the local spin density in real space, but using this information it would be possible to construct a Fourier transform into energy/momentum space to construct the spectral function {$S(\omega, k)$}. We will do this calculation for a finite size system. A better approach would be to use 'infinite boundaries'; that is, an excitation that exists as a finite window in the middle of a translationally invariant iMPS (see Phys. Rev. B 86, 245107 for details), however the To construct a finite size state, we can re-use the lattice files and operators for an infinite system. So we start with a spinchain with {$U(1)$} symmetry,
Now to use finite-size DMRG we need an initial state. There are several ways to construct this initial state, but one of the simplest is to start from a random state.
This constructs a random state with 100 sites, with quantum number 0 (being the z-component of spin). We can now use this as input to the finite DMRG program, mp-dmrg -w psi -H lattice:H_J1 -m 10..100x10,100x10 mp-dmrg -w psi -H lattice:H_J1 -m 100x10 --mix-factor 1e-4 mp-dmrg -w psi -H lattice:H_J1 -m 100x10 --mix-factor 1e-6 In this calculation, we started with a basis size of m=10 states, ramping up to 100 states over 10 sweeps. We then performed another 10 sweeps with the density matrix mixing factor reduced to 1e-4, and another 10 sweeps with mixing factor 1e-6. This produces a quite well converged wavefunction; we could keep going with smaller mixing factors and more sweeps but it won't change much in the wavefunction (increasing the basis size would have the single biggest effect on the accuracy). We can now construct a local spin excitation, by acting on the wavefunction with an operator, for example {$S^+$} acting on the 50th site.
We can view the local spin density of the excitation using gnuplot:
This produces output similar to: 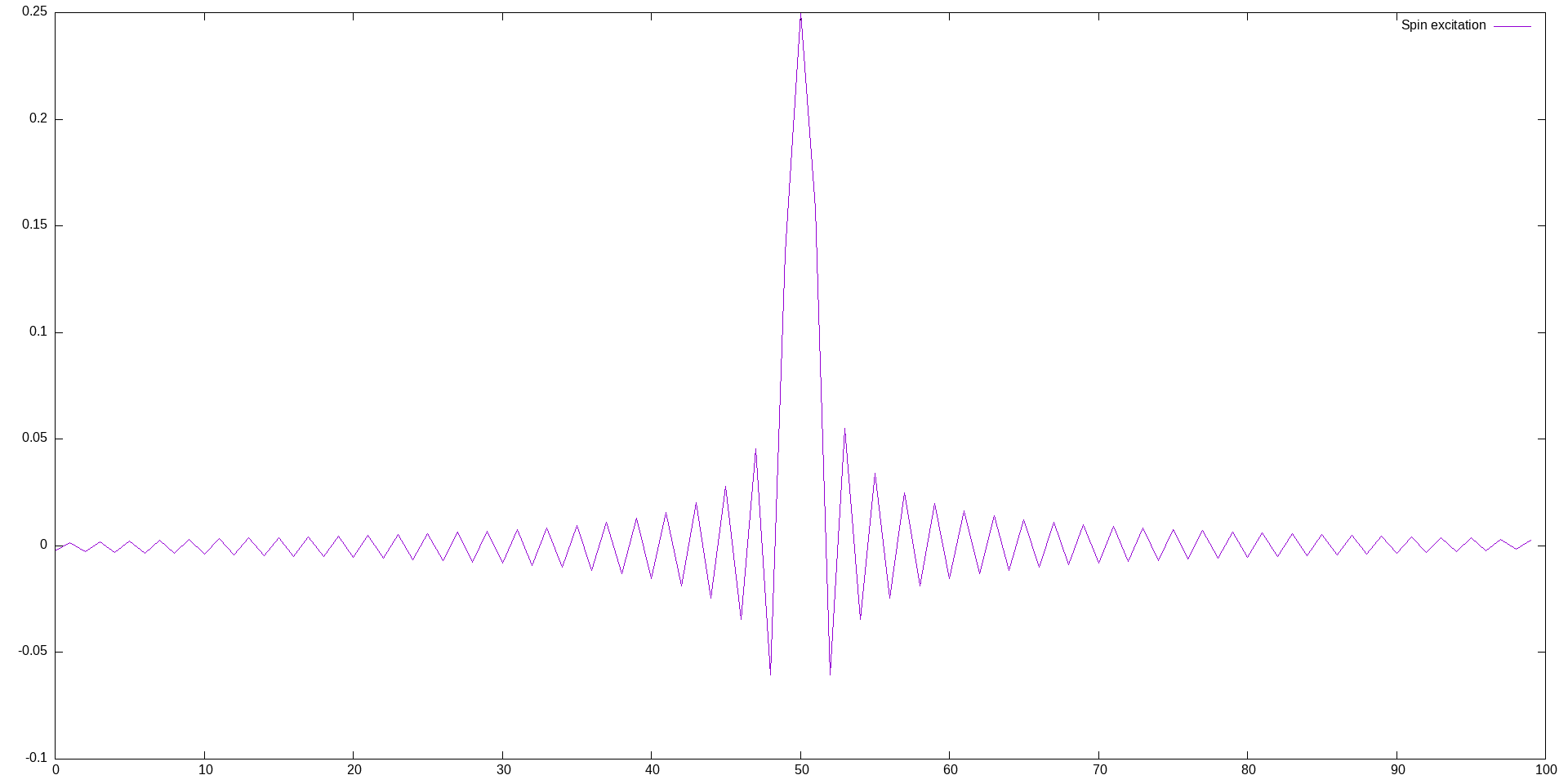 It is slightly asymmetric, because the open boundary conditions induces a slight odd/even dimerization, but this doesn't matter for our purposes. Now we can evolve the state in real time using TEBD. To see the various options of the
The We didn't specify the Hamiltonian here; this will default to the Hamiltonian used to construct the wavefunction, but if we wanted to specify a different Hamiltonian then we could supply one with the usual This will take a few minutes to run (about 5 minutes, on my laptop), and will produce 200 wavefunctions, one for each timestep from 0.1 up to 20.0. Now we can calculate the local magnetization for each wavefunction. We could probably do this directly from Gnuplot, but it is annoying to have plot commands that take a long time to run, so we will save the data to a file using a bash script. cp psi-excitation psi-evolved.t0.0 for t in $(seq 0.0 0.1 20.0) ; do ( for x in $(seq 0 99) ; do echo -n $x ' ' ; mp-expectation --real psi-evolved.t$t lattice:"Sz($x)" ; done > magnetization-t$t.dat ) ; done Note that the first thing that we did was copy the t=0 initial state to the file psi-evolved.t0.0, so that we have a consistent naming scheme. Then our bash loop iterates over each file, writes a data file named Now we can plot the result using Gnuplot: gnuplot> set xrange [20:80]
splot for [t=1:99] 'magnetization-t'.sprintf("%.1f",t/10.0).'.dat' u 1:(t):2:2:2 w lines palette lw 6 title ''
The output should look similar to: 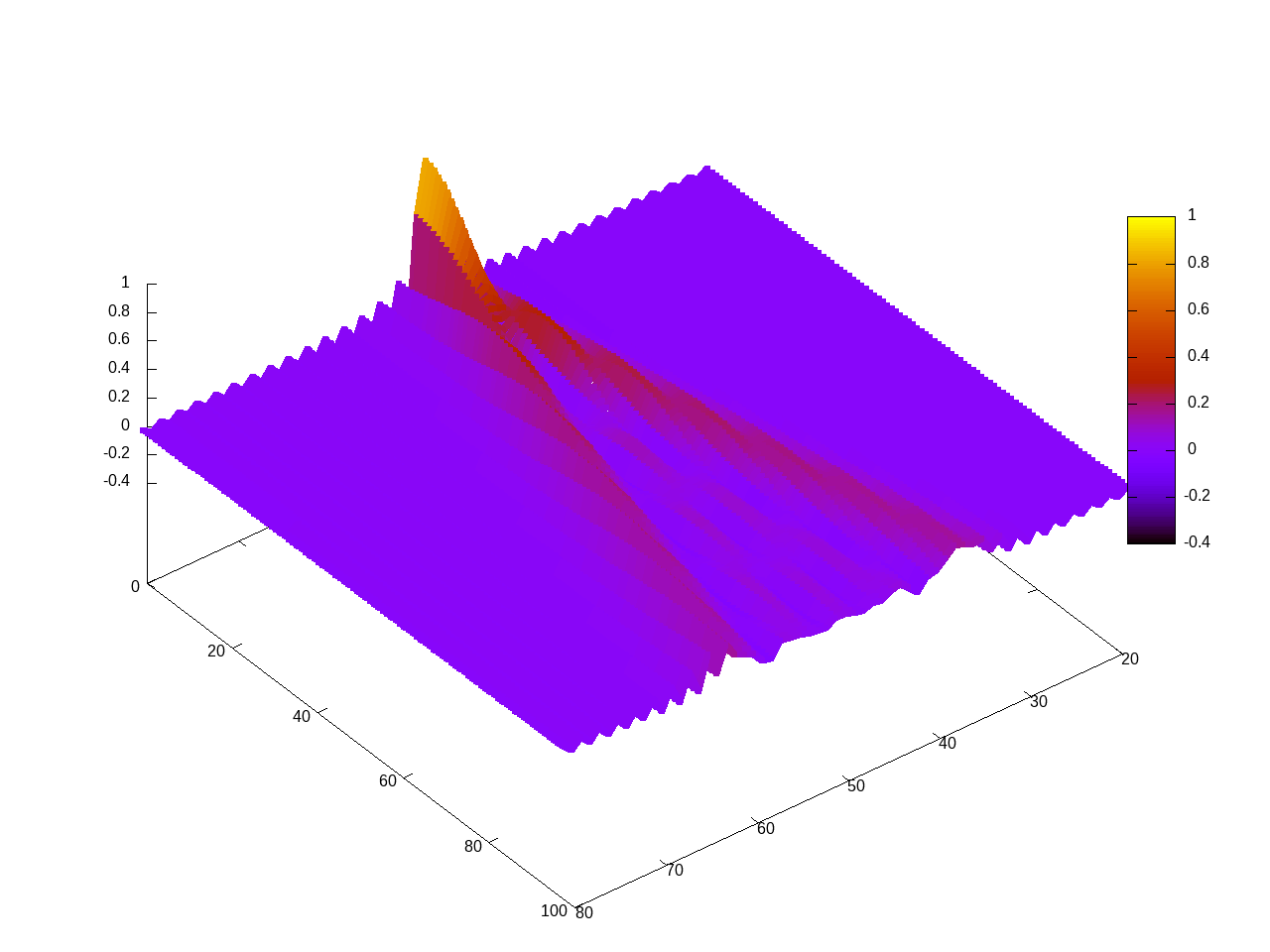 |